 ANSI/HL7 V3 ECG,R1-2004 (R2009) HL7 Version 3 Standard: Regulated Studies - Annotated ECG, Release 1 8/27/2009 |
 HL7 V3 IG ECG, R1-2004 HL7 Version 3 Implementation Guide: Annotated ECG, Release 1 2004 |
Content Last Edited: 2012-08-06T15:01:40
2.2 Storyboards
2.3 Application Roles
2.4 Refined Message Information Models
2.5 Hierarchical Message Descriptions
2.A Implementation Guide
Annotated ECG (aECG), Release 1 is an HL7 and ANSI approved standard..
The Implementation Guide for aECG, Release 1 has also been approved as an HL7 Informative Document. You can download the implementation guide from the Annex at the end of this topic.
The aECG standard is presented here for reference and implementation.
aECG Background
Clinical trials on candidate drug products often collect biological data from trial subjects as waveforms or algorithmic representations of those waveforms. After the data have been collected, derived measurements such as QT interval are used to draw conclusions about the safety and effectiveness of the candidate drug. Standards exist for transmitting these derived data and their analysis results from the trial sponsor to the regulatory agencies; but these submissions have not historically included representation of the waveforms, or have at best included an image of the device-generated paper waveform. A new request to include the digital waveform, the algorithmic representations, and the annotations identifying the relevant landmarks (e.g. QRS-wave onset, T-wave offset) that support derived ECG measurements has been proposed by the US FDA. This specification will be used to package such annotated digital waveform data produced by an ECG analysis system for transmission from trial sponsor to a regulatory agency. A medical device that acquires waveforms directly from patients for presentation to caregivers in support of patient care (e.g., ECG monitors, 12 lead ECG carts, ambulatory ECG recorders, pulse oximeters, etc.) may, but is not required to, comply with this specification.
Limitations on the Annotated ECG Standard
Application of this Annotated ECG message is limited to human subjects only. At this time, no provisions have been made to capture information about animals such as breed and species.
There is no capability to break down the trial into phases, parts, arms, etc. in this message. Therefore, treatment assignments and study events all apply to the trial as a whole.
This message is intended to interchange annotated ECG waveforms recorded only from the person designated as the trial subject.
Future extensions may include the ability to reference non-human subjects, to specify subparts of trials, and to collect waveforms from individuals of interest other than trial subjects.
|
||||||
|
For details on the interpretation of this section, see the storyboard discussion in the Version 3 Guide.
The Storyboard is presented as a high-level description of the business situation the standard is designed to address, accompanied by a diagram of the interactions that will be described in the narratives that follow. Interactions were discussed in the Introduction and Scope section of the Regulated Studies domain chapter, under the heading "Summary of Clinical Trial Interactions in the Regulated Studies Domain". There you will find a diagram of the AnnotatedECG and Clinical Trial laboratory interactions in context.
A number of narrative sections follow each high-level description, each describing a scenario in which one of the interactions might be used. These narratives are for illustrative purposes only. They are not meant to limit the applicability of the standard, nor to proscribe minimum requirements for using the standard. In particular, there is no intent to establish regulatory agency policy on management of data or processes through these scenarios. Such policy will be separately published by the regulatory agencies (eg. as Implementation Guides or Guidances).
The purpose of this storyboard is to illustrate a typical process of collecting, analyzing, and annotating ECGs taken during a clinical trial, and submitting the annotated ECG waveforms to a regulatory agency.
The following diagram illustrates the overall workflow for this storyboard.
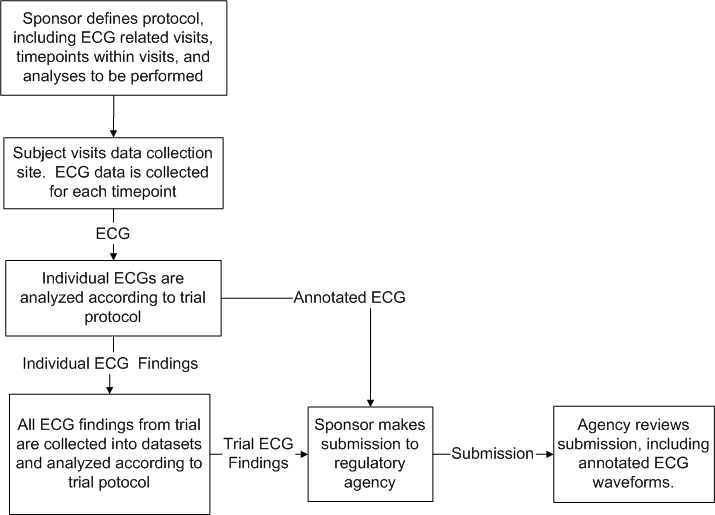
The Storyboard Narratives that follow illustrate the interactions pictured below. It is important to understand that the interaction "AnnotatedECGRevised, Notification" includes creation of new AnnotatedECGs (revised from non-existence into existence).
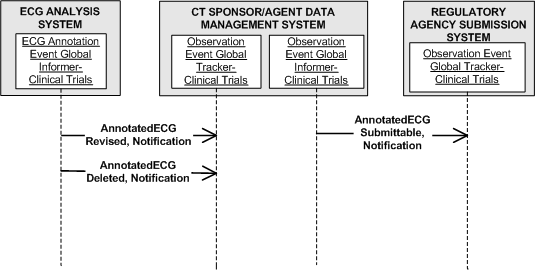
Interactions: AnnotatedECG Revised, Notification (PORT_IN020001) AnnotatedECG Deleted, Notification (PORT_IN020002)
The story starts when the Sponsor decides to study the effects of a new drug on a population's cardiac electrophysiology. The sponsor decides the effects to be studied can be seen in ECG data, or information derived from ECG data. The sponsor designs a protocol specifying the population to be studied, the data to be collected, the equipment to be used for gathering the ECG data, the analysis tools to use, and how any effects will be studied using the ECG data and analysis tools.
The sponsor (or a Contract Research Organization (CRO) engaged by the sponsor) hires investigators to collect the data, and the investigators in turn enroll subjects in the trial. The subject visits the collection site. During the subject's visit, equipment is connected to the subject. The equipment records the subject's ECG along with other physiologic parameters, if appropriate. Trial and subject identifying information is associated with the ECG data, such as subject id, gender, date of birth, investigator and site id's, protocol id, etc. After the individual ECGs are collected from the subject for that visit, it is transferred to a central ECG lab for analysis.
The ECG waveforms can be loaded into an analysis tool for making findings (measurements) and possibly deriving other data according to the trial protocol. The findings are oftentimes in the form of time and amplitude values for the parts of the cardiac process being studied. The analysis produces two types of information: findings to be used for further statistical analysis on the entire population enrolled in the trial, and annotated ECG waveforms with trial specific fiducial markings, etc. indicating key features used for obtaining the findings. The annotated ECG waveforms are forwarded to the trial sponsor (interaction: AnnotatedECG Revised, Notification (PORT_IN020001)), who can include them without ammendment in a trial submission, or may supply some additional data not present in the notification from the central ECG lab. In the latter case, the sponsor is responsible for maintaining an audit trail of changes made to the message as received.
Should any errors be discovered in the message after notification, the deletion interaction (interaction: AnnotatedECG Deleted, Notification (PORT_IN020002)) is issued by the central ECG lab, and a new notification message with the corrected information is issued (interaction: AnnotatedECG Revised, Notification (PORT_IN020001)).
Interaction: AnnotatedECG Submittable, Notification (PORT_IN020003)
The supporting annotated ECG waveforms become part of the sponsor's submission to the regulatory agency. The communication of the annotated ECG waveforms from the sponsor to the regulatory agency (interaction: AnnotatedECG Submittable, Notification (PORT_IN020003)) is the primary focus of this standard.
During the regulatory agency's review of a submission, the Agency may come across a conclusion drawn from a statistical analysis of ECG findings it wishes to investigate further. The reviewer may review the ECG analysis protocol and the analysis dataset. The reviewer may question an ECG finding in the dataset and review the annotated ECG waveforms. The reviewer may be satisfied with what he sees or may investigate further the ECG measurement protocol and its consistent application during the trial.
Data Describing the Clinical Trial Context of the Annotated ECG
Clinical Trial Context Diagram for Annotated ECGs: 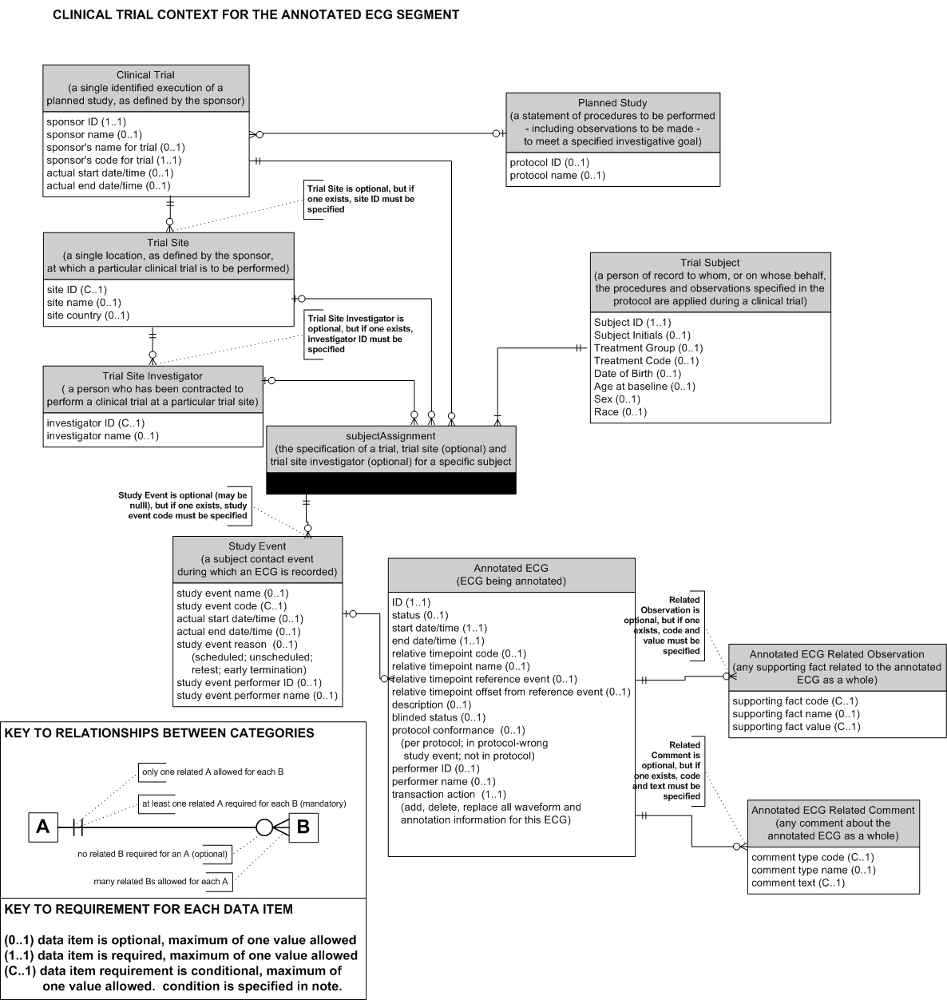
The annotated ECG waveforms are the focus of this message. For each annotated ECG, information is included describing the trial conditions under which the ECG was recorded. At a minimum, the unique identifiers for the ECG, the trial subject, the trial itself are required. In addition to the required identifiers, the message must contain the time of the ECG data collection. This is the minimal set of data considered necessary to properly identify an ECG within a regulatory submission.
These requirements are included in the diagram above, which shows the annotated ECG itself as a category of data to be included in the message. Other trial context categories are also shown, along with the relationships among the categories. All of the individual trial context data items that can be communicated using the HL7 Annotated ECG message are listed and categorized on the diagram, with an indication of whether they are optional or required. Some entity-relationship (ER) constructs and conventions have been used in this diagram, as described in the key, but this is not intended to be a formal ER diagram.
Data Describing the Annotations and Waveforms
ECG Data Breakdown Diagram for Annotated ECGs: 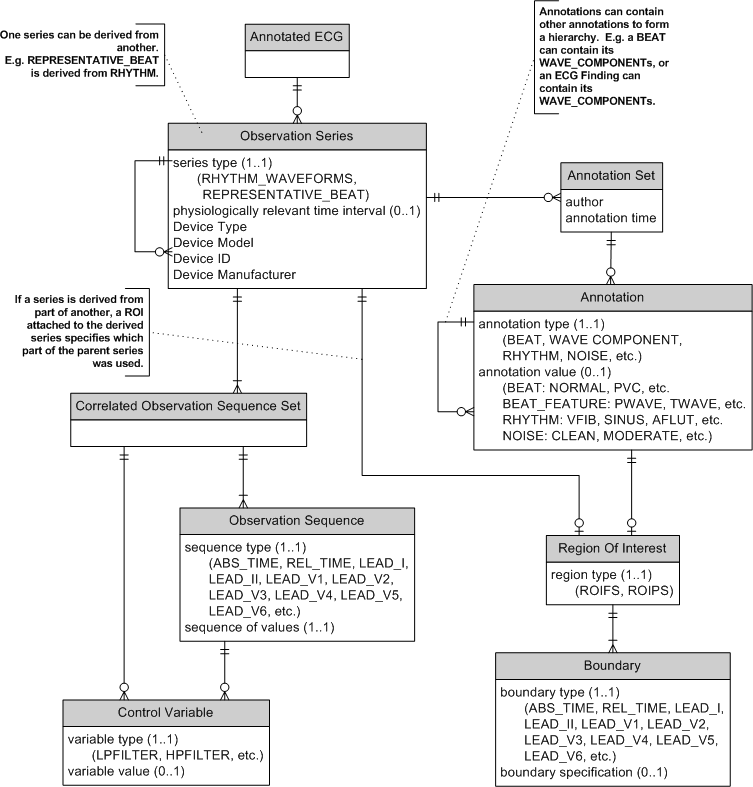
The diagram above shows how the waveforms and annotations are organized relative to the main Annotated ECG object from the previous diagram. Starting from the bottom of the diagram, each lead and the time associated with the samples is represented as a Sequence -- a sequence of values. Each Sequence has a type (lead name or time type) and the list of values.
Sequences that were recorded simultaneously are grouped as Sequence Sets. A modern electrocardiograph can represent a 12-lead ECG as a single Sequence Set. A 3 channel electrocardiograph would typically use 4 Sequence Sets for a 12-lead ECG (e.g. set 1 having leads I, II, III, set 2 having leads aVR, aVF, aVL, set 3 having leads V1, V2, V3, and set 4 having leads V4, V5, V5).
A Series contains all the Sequence Sets that share a common frame of reference. Typically, the Series will contain the data collected from a single device with a single time reference. If a representative beat is algorithmically derived from the rhythm data, it will form another Series (derived from the rhythm data Series).
A number of Annotation Sets can be attached to the Series. The Annotation Sets contain an arbitrary nesting of Annotations. The nesting of Annotations allows parent/child relationships to be communicated. For example, a Beat annotation can be the parent of Wave Component annotations (P-wave, QRS-wave, T-wave, etc.).
If an annotation has a location within the Series, it is associated with a Region Of Interest (ROI). The ROI specifies the leads and time period for the annotation. A fully specified ROI contains a Boundary for each lead and/or time dimension within it. A partially specified ROI will contain a Boundary for each lead and/or time that is not wholly in it. For example, if a global QRS onset is to be specified, a partially specified ROI will only have a Boundary for the time dimension. It is assumed that all the other dimensions (leads) are wholly in the ROI.
|
||||||
|
For details on the interpretation of this section, see the discussion of application roles and their relationships in the Version 3 Guide.
The AnnotatedECG topic has three application roles, two of which are shared with the CTLaboratory topic in this domain.
Those shared application roles are defined under the CTLaboratory topic. Click on their names below to see those definitions:
Clinical Trial Observation Event Global Tracker
Clinical Trial Observation Event Global Informer
An application that is capable of notifying another system about any state change of a Clinical Trial Annotated ECG Event.
|
||||||
|
For details on the interpretation of this section, see the description of RMIMs in the Version 3 Guide.
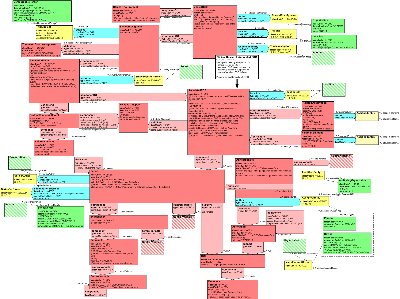
| Parent: | None |
The Refined Message Information Model (RMIM) recasts the committee's business models in terms of the HL7 Reference Information Model (RIM). Click on the links below to open diagrams of those business models.
Business Model - Clinical Trial Context:
Business Model - ECG Data Breakdown:

It will be helpful to open the RMIM at this time to follow along with the discussion of how the RMIM represents the business requirements.
REPRESENTATION IN THE R-MIM OF THE INFORMATION ABOUT CLINICAL TRIAL CONTEXT
Business Model Category: Annotated ECG
ID, start/end datetimes, blinded status, and protocol conformance specified in the business model are all represented in the R-MIM class AnnotatedECG, which appears in the approximate center of the RMIM diagram. Note that start and end date and time are all captured in the effectiveTime attribute, which can represent an interval of dateTimes.
The relative timepoint information for the ECG is represented in the R-MIM classes ProtocolTimepointEvent, ReferenceEvent and RelativeTimepoint. The ProtocolTimepointEvent is the protocol definition of the TimepointEvent during which the ECG is taken. If the protocol does not include the notion of timepoint or study events, then the entire trial is considered a single timepoint event for purposes of establishing the ReferenceEvent and RelativeTimepoint.
The ReferenceEvent class corresponds to the benchmark for the ECG within the TimepointEvent. This could be a dosing for example, or a meal or any other benchmark to which the ECG is related. The protocol-specified name and code (if any) for the benchmark are captured in the code attribute of this Reference Event class.
The RelativeTimepoint class represents the (scheduled) time for the specific ECG relative to the Reference Event, and the offset value (positive or negative amount and UOM for the offset of the ECG from the benchmark) is captured in the pauseQuantity attribute of the componentOf relationship between the ProtocolTimepointEvent and the RelativeTimepoint classes.
Performer information for each ECG Series is captured in the R-MIM's ECGPerformer role and its related Person.
The Transaction Action information is captured in the message wrapper (not shown here).
Business Model Category: Annotated ECG Related Observation
All business information in this category is captured in the R-MIM's RelatedObservation class, where the code identifies the type and name for the observation (eg. age, fasting status) and the value captures the observation itself.
Business Model Category: Annotated ECG Related Comment
All business information in this category is captured in the R-MIM's FindingComment class, where the code identifies the type and name for the comment and the value captures the comment itself.
Business Model Category: Study Event
All of the business information in this category is captured in the TimepointEvent class of the R-MIM, except performer information, which is represented by the StudyEventPerformer class and its related Person.
Each AnnotatedECG must be assigned to exactly one TimepointEvent. If the protocol does not include the notion of timepoint or study events, then the entire trial is considered a single timepoint event.
Business Model Categories: Clinical Trial; Trial Site; Trial Site Investigator
The Clinical Trial category in the business model has a counterpart ClinicalTrial class in the R-MIM. This class captures the sponsor's name and code for the trial (not the protocol) in the id and title attributes. The activityTime attribute captures the start and end dates (and times, if necessary) for the entire trial.
The name and identifier for the sponsor is represented in the Organization class, which plays the role of ClinicalTrialSponsor, and participates in the trial as the trial's author.
Trial site information (id, name, country) is captured in the TrialSite role class, or in the Location entity class which plays that role.
Similarly, trial investigator information (id, name) is captured in the TrialInvestigator and Person classes. The identifier of the person as an investigator should appear in the TrialInvestigator role id.
Business Model Category: Planned Study
Planned Study in the business model represents the protocol controlling the individual clinical trial. It is possible for several individual clinical trials to be controlled by the same protocol. The id and name for the controlling protocol for this specific trial are captured in the ClinicalTrialProtocol class of the R-MIM.
Business Model Categories: Trial Subject; Subject Assignment
Most of the trial subject information is captured in the R-MIM classes TrialSubject (subject id) and DemographicPerson (initials, birthdate, gender, race).
As in the business model, the R-MIM class SubjectAssignment serves to associate the subject with a particular trial, site and investigator. Assignment to a trial is mandatory; assignment to a site and investigator are optional.
In the R-MIM, the SubjectAssignment class also serves to associate the subject with the treatment group to which he/she is assigned for the trial.
REPRESENTATION IN THE R-MIM OF THE INFORMATION ABOUT WAVEFORMS AND ANNOTATIONS
The R-MIM class Series contains the rhythm waveforms directly recorded from the subject, or the representative beat waveforms derived from the rhythm waveforms. The code attribute indicates if the series contains rhythm waveforms or representative beat waveforms.
The Series attribute effectiveTime captures the interval of time for the waveforms (usually recorded by the device). The effectiveTime for rhythm waveforms is the actual time of data collection. The effectiveTime for a representative beat is the period of time from which the beat was derived.
The Series can have a unique id, usually assigned by the device that authored it. This unique id can be different from the id of the AnnotatedECG because there can potentially be several Series contributing to a single AnnotatedECG (for a single TimepointEvent and RelativeTimepoint). The Series id also allows annotations in other messages to be associated with the Series.
The Series is authored by a Device playing a ManufacturedProduct role. The role is scoped by the Organization that manufactured the device. The Device entity captures the id, code (type), model name, and software name. The Organization entity captures the manufacturer's id and name.
If necessary, id's and names (or initials) of one or more technicians acquiring the ECG data can be captured as the Series SecondaryPerformer. The SecondaryPerformer is commonly referred to as the "ECG Technician".
If the Series is a representative beat, and it was derived from a portion of the rhythm series, it is supported by an ROI (Region Of Interest). The ROI specifies which segment of time the representative beat was derived from.
The Series has one or more SequenceSet components. There is one SequenceSet for each set of ECG leads that were simultaneously acquired. A modern electrocardiograph can simultaneously capture all 12 leads of a 12-lead ECG, and therefore only requires a single SequenceSet. A 3-channel electrocardiograph, on the other hand, must acquire the 12 leads 3 at a time. A 3-channel device would typically use 4 SequenceSets for a 12-lead ECG.
Each SequenceSet contains a Sequence for the time base and each lead simultaneously acquired. For example, a 12-lead ECG represented in a single SequenceSet would have 13 Sequences: one for time, and one for each of the 12 leads. The type of Sequence is captured in the code attribute, and the sample values are captured in the value attribute (as a list). The ECG lead types from the ISO/IEEE 11073-10101 standard will be used for the code attribute whenever appropriate.
It is important to note that HL7 provides a couple of special datatypes for representing the values of a Sequence. If the Sequence contains a regular pattern of values (such as evenly sampled time), the GLIST (generated list) collection type can generate the sequence of values from a few parameters (e.g. initial value and increment). If the Sequence contains values in native integer device units, the SLIST (sampled list) collection type can specify scale and translate factors so the native integers can be placed in the message
The Series, SequenceSet, and/or Sequence can be associated with one or more ControlVariables. A ControlVariable captures additional information about how the waveforms were created. For example, if the waveforms had filtering applied, the filter settings should be captured as ControlVariables. The ControlVariable code captures the type of variable (e.g. low pass filter), and the value captures the variable's setting (e.g. 150 Hz).
AnnotationSets can be associated with a Series. The activityTime of the set captures when the set of annotations were made. Each set of annotations can have a common author and common ControlVariables. The author can be a Device or a Person playing the AssignedEntity role. The id of the AssignedEntity captures the trial specific id of the annotator. The scoping Organization captures the id and name of the organization making the annotation via the Device or Person. ControlVariables can be associated with the AnnotationSet to capture important details about the annotation method (e.g. algorithm settings such as criteria used to classify tachycardia, etc.).
Each AnnotationSet contains one or more annotations for the Series. Each Annotation has a code specifying what type of annotation it is, and when appropriate, the value attribute specifies the annotations's value. The ECG nomenclature from the ISO/IEEE 11073-10101 standard will be used for the Annotation codes and values whenever possible.
Annotations can contain child annotations when appropriate. For example, it may be necessary to communicate that a P-wave, QRS-wave, and T-wave all belong to the same beat. A beat annotation could then contain child annotations for each of the wave components. Likewise, it might be necessary to group all the fiducial markings related to a particular ECG measurement, like QTc. A QTc annotation could have child annotations for a couple R-peaks, a QRS-wave, and a T-wave. If the QTc was calculated from several beats, each beat's QTc fiducial markings could make up another level in the hierarchy.
Each Annotation can be supported by an ROI. The ROI specifies where in the Series the Annotation applies. For example, if the Annotation is a T-wave, the ROI will specify a Boundary in time for the beginning and end of the T-wave, and optionally a Boundary for each Lead that is included.
The ROI code attribute says whether the ROI is partially or fully specified by its Boundaries. A partially specified ROI only include Boundaries for dimensions that are constrained. All other unspecified dimensions would be included in the ROI. For example, a global QRS-wave could be annotated with a partially specified ROI with only a time Boundary. A fully specified ROI, on the other hand, only includes the dimensions that are named as Boundaries. So, if the QRS-wave is only observed in Lead II, then a Boundary for time and a Boundary for Lead II would be specified. All other unnamed dimensions would not be included in the ROI.
The code attribute of the Boundary names the dimension being bounded. These codes should match the codes used in the Sequence instances. For example, if the Series contains a Sequence for absolute time, and an ROI needs to bound that time, the same code for absolute time is used as the Boundary code. The Boundary's value attribute is usually an interval capturing the optional start and optional end of the ROI within that dimension. However, if the value is truly a single point (e.g. R-wave peak), then the value attribute will contain a single value, not an interval.
The Boundary values can be used to communicate several things. If a T-wave Annotation is used to communicate the offset of T, the ROI's time boundary would only specify the interval's high value. The low value can be unspecified. Likewise, if the beginning of the a P-wave is to be communicated, the ROI time boundary would only specify the interval's low value.
The relationship between the Annotation and its ROI can also be used to communicate a couple of different concepts. If the relationship is the more general "has support", the ROI communicates boundaries that aren't necessarily the true boundaries of the annotation. For example, if atrial flutter is observed in the waveforms, but the true beginning and ending of the flutter episode is not observed, the annotation "has support" from the ROI (but it's not the full story). On the other hand, if the true beginning and end of atrial flutter is observed, the "has bounded support" relationship can be used. In other words, the annotation's ROI supports AND defines the true boundaries.
| AnnotatedECG - Clinical Trials | PORT_HD020001UV01 |
|
||||||
|
For details on the interpretation of this section, see the description of HMDs in the Version 3 Guide.
The AnnotatedECG - Clinical Trials HMD is based upon the Annotated ECG - Clinical Trials R-MIM. All information available from that RMIM is included in this HMD. Message types based upon this HMD are used to communicate ECG annotations and waveforms.
| AnnotatedECG. base - Clinical Trials | PORT_MT020001UV01 | |
| AnnotatedECG, deletion - Clinical Trials | PORT_MT020002UV01 |
This annex presents the HL7 implementation guide for the approved standard Annotated ECG, Release 1. The guide is a stand-alone document.
Click here to view a PDF of the approved implementation guide.
| Return to top of page |

