 ANSI/HL7 V3 CGPED, R1-2007 (R2012) HL7 Version 3 Standard: Clinical Genomics; Pedigree, Release 1 (reaffirmation of ANSI/HL7 V3 CGPED, R1-2007) 6/21/2012 |
Content Last Edited: 2012-02-24T15:12:20
2.2 Application Roles
2.3 Trigger Events
2.4 Refined Message Information Models
2.5 Hierarchical Message Descriptions
2.6 Interactions
|
||||||
|
For details on the interpretation of this section, see the storyboard discussion in the Version 3 Guide.
The objective of this storyboard is to illustrate the way a patient's pedigree with clinical and genomic data could be represented for risk analysis purposes in the context of breast and ovarian cancers and other diseases. The initial context for this storyboard was set by the BRCA storyboard which is still under development. The UML Activity Diagram below as well as the Presentation section further on are taken from the BRCA storyboard, illustrating the use of family history data in this case.
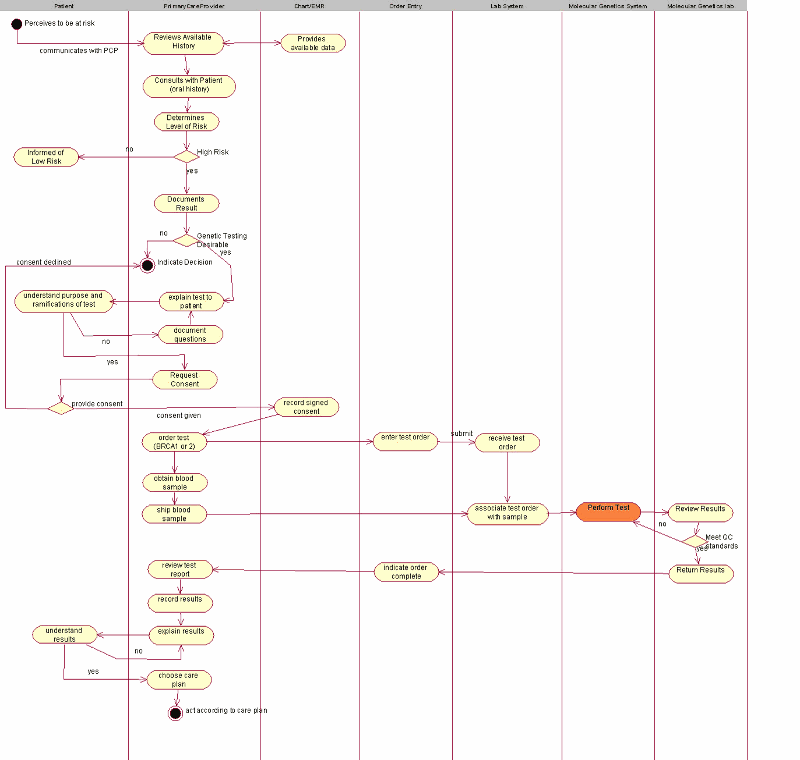
| Original Pedigree |
Summary
The Family History storyboard and its HL7 Pedigree Model is part of the HL7 Clinical-Genomics SIG effort to design various higher-level models of using genomic data in healthcare practice, that utilize the basic model of a Genotype (described at this point as the Clinical Genomics Domain Information Model). The need to represent a patient's pedigree information as associated with clinical and genomic data was introduced in the BRCA (Breast Cancer) storyboard which is still under development.
The BRCA storyboard includes a sample outline for the way the patient's pedigree should look:
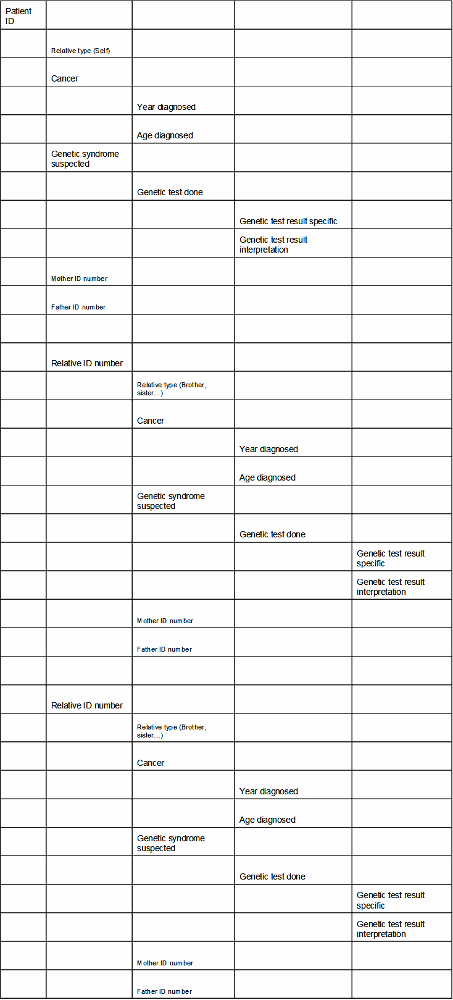
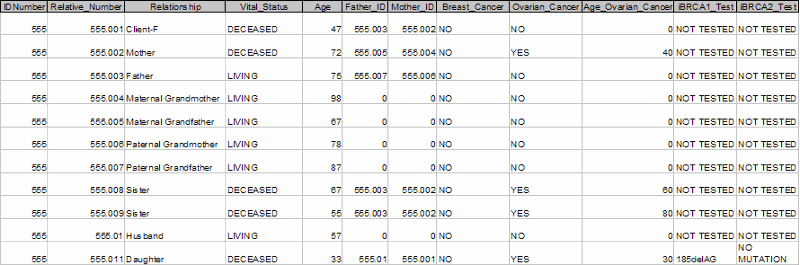
Populating the above outline with actual data might result in a spreadsheet found in the following link:
Presentation
- Ms. Eve Everywoman has a family history of breast and ovarian cancer, and she is not of Ashkenazi Jewish descent.
- She believes she is at high risk of developing breast cancer.
- She goes to see her clinician (Medical oncologist, surgical oncologist , radiation oncologist, primary care provider) who takes a thorough family history. This history is recorded in the chart and the electronic medical record.
- The clinician reviews the family history, decides what genetic syndrome her family might have, and categorizes the patient as to degree of risk (Perhaps high, medium, or low risk). The clinician thinks the patient is at high risk of having a BRCA1/2 mutation.
- The clinician compares her Family History to tables of risk (Claus, Myriad) and runs computer models (algorithms such as BRCAPRO, see http://www.isds.duke.edu/~gp/brcapro.html). This gives a percentage risk of carrying a mutation and/or a risk of developing breast and/or ovarian cancer. Her risk of a mutation is 25%, because her father's 4 sisters had ovarian caner.
- The patient is considered to be at high risk of having a mutation, and this information is given to her.
- She is referred to a Risk Clinic.
- She agrees to go to the Risk Clinic.
- Ms Eve Everywoman's Genetic History details are sent to this clinic (the HL7 Interaction POCG_IN000001 is used), including her Family History, the syndrome suspected and her level of risk.
- The counselor at the risk clinic (Nurse geneticist, genetic counselor, MD, etc.) reviews the family history information collected by the primary clinician, edits it and adds additional details.
- The Counselor reviews the family history, decides what genetic syndrome her family might have, and categorizes the patient as to degree of risk (Perhaps high, medium, or low risk). The Counselor thinks the patient is at high risk of having a BRCA1/2 mutation.
- The clinician compares the family history to tables of risk (Claus, Myriad) and runs computer models (algorithms such as BRCAPRO). This gives a percentage risk of carrying a mutation and/or a risk of developing breast and/or ovarian cancer. Her risk of a mutation is 25%, because her father's 4 sisters had ovarian caner.
- The patient is considered to be at high risk, and she is told she is a candidate for genetic testing. This includes a thorough discussion of the pros and cons of testing. This discussion is recorded in the electronic medical record.
- Ms. Eve Everywoman wants to have testing, but as she is not affected, it is the standard of care to test a living affected relative first.
- The Counselor suggests that her Aunt, Ms. Jeanne Aunt, is the most appropriate candidate for testing. Ms. Jeanne Aunt had ovarian caner, and is still living.
- Ms. Eve Everywoman agrees to contact Ms. Jeanne Aunt.
- Ms Eve Everywoman signs consent to release her own Family History details to Ms Jeanne Aunt and her Provider.
- Ms. Jeanne Aunt is a 39-year-old woman had been diagnosed with ovarian cancer at age 35.
- Ms Jeanne Aunt agrees to discuss testing, and provides the name and address of the Risk Clinic she will attend.
- Ms Eve Everywoman's FH details are sent to this clinic (the HL7 Interaction POCG_IN000001 is used).
- The counselor at the risk clinic (Nurse geneticist, genetic counselor, MD, etc.) reviews the family history information collected by the primary clinician through a pedigree drawing program, and changes the Proband* to Ms Jeanne Aunt, edits it and adds additional details (The family history message had had Ms Eve Everywoman as the Proband (Self), and Ms Jeanne Aunt as the aunt. The pedigree from the point of view of Ms Jeanne Aunt must have Jeanne Aunt as the Proband (Self) and must show Ms Eve Everywoman as the niece).
- The Counselor reviews the family history, decides what genetic syndrome her family might have, and categorizes the patient as to degree of risk (Perhaps high, medium, or low risk). The Counselor thinks the patient is at high risk of having a BRCA1/2 mutation.
- The clinician compares the family history to tables of risk (Claus, Myriad) and runs computer models (algorithms such as BRCAPRO). This gives a percentage risk of carrying a mutation and/or a risk of developing breast and/or ovarian cancer. Ms. Jeanne Aunt is virtually at 100% risk of having a mutation.
- The patient is considered to be at high risk, and she is told she is a candidate for genetic testing. This includes a thorough discussion of the pros and cons of testing. This discussion is reviewed in the electronic medical record.
- Ms. Jeanne Aunt wants to have testing. She signs an informed consent document.
- The order for testing is issued, and the informed consent, and the family history are included with the lab requisition. All are MESSEGED to the blood drawing facility. In case the family history is messaged separately, then HL7 Interaction POCG_IN000001 is used.
- The blood is drawn, and sent to the central testing facility along with the informed consent, the family history, the results of Ms Jeanne Aunt's test, and the lab requisition.
- At the central testing facility, the specimen is checked in, and the DNA is separated and PCRed.
- Full gene sequencing of BRCA1 and BRCA 2 are undertaken.
- The sequence is assessed for mutations.
- Identified mutations are assessed for functional significance by determining if they are truncating (deleterious), or if they are irrelevant (No change in amino acid coded by that codon). All other mutations are compared to known mutations to determine if information is available on their functional significance.
- The actual mutation, and the assessment of functional significance are sent to the counselor.
- In this case, a mutation is identified in BRCA1 and the mutation is Deleterious.
- The counselor discusses the result with the patient.
- Management decisions (Screening, chemoprevention, prophylactic surgery) are probably beyond the scope of this storyboard.
- Ms Jeanne Aunt agrees to share this information with Ms Eve Everywoman's Clinician.
- This information is sent to Ms Eve Everywoman's Clinician.
- Ms. Eve Everywoman wants to have testing. She signs an informed consent document.
- The order for testing is issued, and the informed consent, and the family history are included with the requisition, as well as the results of Ms Jeanne Aunt's test. All are MESSEGED to the blood drawing facility.
- The blood is drawn, and sent to the central testing facility along with the informed consent, the family history and the lab requisition.
- At the central testing facility, the specimen is checked in, and the DNA is separated and PCRed.
- Full gene sequencing is not needed. Testing only for the identified mutation is undertaken.
- The DNA is assessed for that specific mutation.
- The mutation is not found.
- The normal result is sent to the counselor.
- The counselor discusses the result with the patient.
- Management decisions (Screening, chemoprevention, prophylactic surgery) are probably beyond the scope of this storyboard.
* Glossary:
Proband: First affected family member coming to medlcal attention.
Consultand: Individual (s) seeking genetic counseling/tetsing.
These definitions are based on: Bennett RL, Steinhaus KA, Uhrich SB, O'Sullivan CK, Resta RG, Lochner-Doyle D, Markel DS, Vincent V, Hamanishi J. Recommendations for standardized human pedigree nomenclature. Pedigree Standardization Task Force of the National Society of Genetic Counselors. Am J Hum Genet. 1995 Mar;56(3):745-52.
The Family History Shared Model
Following the above sample of a patient's pedigree as well as the contextual presentation, we have developed an HL7 model to allow the representation of such a pedigree with an unlimited depth of generations. This model is described in detail (including a walk-through) in the section: "Refined Message Information Models".
|
||||||||
|
For details on the interpretation of this section, see the discussion of application roles and their relationships in the Version 3 Guide.
|
||||||
|
For details on the interpretation of this section, see the discussion of trigger events in the Version 3 Guide.
|
||||||
|
For details on the interpretation of this section, see the description of RMIMs in the Version 3 Guide.
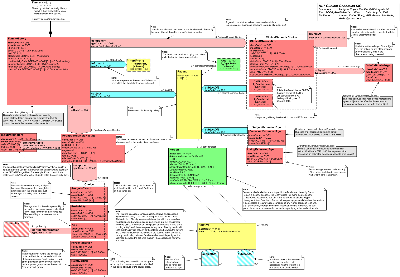
| Parent: | Clinical Genomics (POCG_DM000023UV) |
The Clinical Genomics Family History Model
Following the above sample of a patient's pedigree as well as the contextual presentation, we have developed an HL7 model to allow the representation of such a pedigree with an unlimited depth of generations. The model addresses the Family History requirements and represents a patient's pedigree while making use of the Genotype model for genomic data. Each family member object is represented in relation to another family member who 'scopes' its role and is designated by a code taken from the HL7 vocabulary "RoleCode", domain ="PersonalRelationshipRoleType".
General Notes:
The Family History model is used in a message interaction designed to serve the need of information exchange between two disparate pedigree applications (see section on message interactions). This interaction does not serve other needs like sending genetic lab orders accompanied by family history. For the latter, there is a need to use Lab messages that might use this model as a payload.
The model utilizes the GeneticLocus model (the Genotype Topic) in order to capture genomic data in any resolution needed. For that end, the GeneticLocus model was packaged as an internal CMET and was also moved to the HL7 Common Domains in the V3 Ballot Package. It is utilized in this model as one of the choices in the main Clinical Genomics choice box (see the model walk through below). The model suggests the use of the Clinical Statement shared model (under development in HL7) to represent the clinical data. Meanwhile, it has a generic ClinicalObservation class to hold common clinical data (e.g., problems, diagnoses, reactions to drugs, allergies, etc.).
Model Walk-Through
Starting Point - FamilyHistory:
The starting point of the model is the FamilyHistory Observation class. This class has several associations, one of which is subject participation of the Patient role played by a Person entity. The latter scopes the Relative class which can hold information about the patient's relatives. This constitutes the backbone of the model. In addition, the entry point is associated with risk analysis results and with problems that can not be attributed to specific family members.
Attributes of FamilyHistory:
- id: holds a unique identifier of this family history instance
- code: the code attribute shall hold a code representing Family History data in general, for example: the LOINC code 10157-6, HISTORY OF FAMILY MEMBER DISEASES or any other code that carries similar semantics.
- statusCode: indicates whether the act of family history observation as a whole has completed, still active, etc. (based on the HL7 RIM Act State Machine)
- methodCode: The methodCode holds the identification of the program creating the family history data
Patient & Person:
The Patient role class is the root of the pedigree but in terms of data, it only captures an id assigned by the family history application on behalf of the provider hosting the family history application. Note that the id is optional and so is the class Provider (the scoping entity of the Patient role). Person is the player entity of Patient and holds general information about the person like gender, birth time, deceased indication, etc.
Note that the gender attribute of the Person class is an "administrative gender" and the way to represent genotypic / phenotypic gender is to populate an instance of the clinical observation class in the clinical genomic choice box.
Clinical and Genomic Data:
At the right side of the Patient role there is a 'subjectOf2' participation that associates the patient (as a 'subject of') to a choice box of zero to many clinical statements as well as genomic data represented by the GeneticLocus model. Note that this association is shadowed at the bottom of the model, associated with the Relative class, which represents a role of a patient's relative.
-
ClinicalObservation
The ClinicalObservation class represents any clinical data that is part of the Person clinical history. Currently, the class has the classCode of 'OBS' which means that it is capable of representing only observations. However, the full expression of a clinical statement will be available when this single class will be replaced by the HL7 Clinical Statement model (under development). The Clinical Statement model will provide the 'grammar' of how various discrete acts (observations, procedures, substance administrations, etc.) are associated to a meaningful clinical statement. Nevertheless, in the January 2007 ballot we added a recursive ActRelationship (sourceOf) to the clinical observation to address use cases where a richer clinical statement is need, as introduced to us by early adopters of the model. The addition of this association is in consistent with the Clinical Statement model, so that when eventually this single observation is replaced with the Clinical Statement model or a derivative of it, this current addition is in consistent with it and will not require substantive changes to the family history implementations.
-
DataEstimatedAge
The DataEstimatedAge class is used to hold the estimated age of the subject at the effective time of the observation (e.g, the diagnosis time). The diagnosis is represented by the source observation , i.e., the ClinicalObservation class. It is used due to the absence of an age attribute in all HL7 classes. We have proposed to RIM harmonization the addition of an age attribute to the Act class as well as other classes, but were asked to try and model this piece of information using associated observations like this class. The data type is an interval to allow a range of ages such as in cases when the patient only remembers that the diagnosis was made when the family member was in her forties for example.
The code attribute shall hold a code representing age of subject at the effective time when the source observation was made for that subject. Its purpose is to allow parser applications to understand this class without referring to its name.
-
-
A_GeneticLocus (CMET)
This CMET is the main model we are developing at the HL7 Clinical Genomics SIG. In principle, the GeneticLocus model can hold relevant genomic data in any resolution required. In the BRCA storyboard, it could be information on mutations that the patient's relatives have or full DNA sequences of the patient's genes at stake.
-
A_GeneticLoci (CMET)
This CMET allows the association of data on a set of loci such as genetic test panel results or gene expression assay.
informant:
This class optionally represents the source of information from which this family history was collected.
PedigreeAnalysisResults:
This class represents the results of analysis done to the data captured in the family history pedigree. Use the code attribute to identify the disease or variation for which the probabilities/risks/etc. are calculated. Use the methodCode to hold the algorithm used to analyze the pedigree.
Attributes of PedigreeAnalysisResults:
* code: identifies the disease or variation for which the probabilities/risks/etc. are calculated. Note that this class can be populated as many times as needed, for each clinical condition which is the is the focus of the risk calculations
* negationInd: can be used to represent the fact that there is no risk found for this family history and the clinical condition in the code attribute
* methodCode: holds the type of the algorithm used to analyze the pedigree (e.g., BRCAPRO)
-
risk
The risk association links the FamilyHistory class to the PedigreeAnalysisResults class and represents the risk associated with that family history. The risk Act Relationship is defined in the RIM as follows: "A noteworthy undesired outcome of a patient's condition that is either likely enough to become an issue or is less likely but dangerous enough to be addressed." The patient condition in this model is the patient's family history and the risk is represented through the classes associated with PedigreeAnalysisResults class.
Consequently, the PedigreeAnalysisResults class and the observation classes associated with it are in 'risk' mood (moodCode= RSK) to express the fact that these are not observations that happened, rather they represent a risk associated wit the family history.
-
InputParameters
The controlVariable association links PedigreeAnalysisResults to input parameters used in the analysis like sensitivity and specificity in the BRCAPRO algorithm. For example, if the code attributes holds "sensitivity" then the value attribute holds the sensitivity itself.
-
Choice
The component association links PedigreeAnalysisResults to a choice box that contains several options to represent the actual results:
-
AnalysisResult
This class is a catcher for any analysis that cannot be represented through the other classes in this choice box.
-
Probability
The value holds a probability of having what is represented in the PedigreeAnalysisResults.code attribute (e.g., disease, variation). The code attribute holds a value that indicates that this is a probability observation.
-
PercentageRisk
The value holds a percentage risk of having what is represented in the PedigreeAnalysisResults.code attribute (e.g., disease, variation). The code attribute holds a value that indicates that this is a percentage risk observation.
-
Relative Risk
The value holds a relative risk of having what is represented in the PedigreeAnalysisResults.code attribute (e.g., disease, variation). The code attribute holds a value that indicates that this is a relative risk observation.
-
Age & Probability
The pertinentInformation association links Age to Probability and multiple traversals of the Age class along with Probability can hold pairs of age-probability data for what is represented in the PedigreeAnalysisResults.code attribute (e.g., disease, variation).
The code attribute shall hold a code representing the concept of "Age" in general (e.g., the value 397659008 "Age" in SNOMED CT).
-
Relative:
This refinement of the HL7 Role Class represents a patient's relative and is scoped by the Person entity which plays the Patient role in the first traversal of the model (see further explanation in the "Person and Relative" bullet below). The cardinality of this association is 0..* which allows for the representation of any number of relatives who all relate to the Person who scopes the role. The Relative class has a classCode = "PRS", defined as "links two people in a personal relationship... the character of the relationship must be defined by a PersonalRelationshipRoleType code..." The latter code is defined by the Relative.code attribute whose value set is drawn from the domain "PersonalRelationshipRoleType". Appendix E includes a table that shows the codes of this domain. Using values from this domain it is possible to designate the relation to the patient or to the patient's family member. Thus, in this model it is possible to use, for example, the code GRMTH (grandmother) for Relative associated directly to the patient, or use the code NMTH (mother) for Relative associated to the mother of the patient. This makes the model more flexible.
The basis of this part of the model is in the RIM definition of family member relationships which are based on the relationship between a scoping entity and a role. For example, the code CHILD is defines as "The player of the role is a child of the scoping entity", and the same goes for any type of family relationship. Note that this is valid not only to the relationship between the patient and a relative directly associated with the patient, rather this is true for any relationship between family members on this pedigree, for example, between the patient's mother (the scoper) and her father (the role).
-
SubjectEstimatedAge
This choice box is associated with the Relative class and holds
two classes concerned with estimated ages of the subject as follows:
-
DeceasedEstimatedAge
The DeceasedEstimatedAge class is used to hold the estimated age when the subject died. It is used due to a lack of age attribute in all HL7 classes. We have proposed to RIM harmonization the addition of age attribute to hold the deceased age as well as the subject age at the time a diagnosis was made (see above in the DataEstimatedAge class description), but were asked to try and model this piece of information using associated observations like this class.
-
LivingEstimatedAge
The LivingEstimatedAge class is used to hold the estimated age of a living relative whose birth date is unknown.
- For a living subject, the code shall represent semantics similar to the LOINC code "21611-9" that represents the concept of an estimated age (as opposed to precise age). For a deceased subject, the code shall represent semantics similar to the LOINC code 39016-1 (AGE AT DEATH).
-
subjectOf Shadow
This class is a shadow of the subjectOf2 class associated with Patient and thus also includes all its associated classes. This means that the same clinical and genomic data structures attached to the patient could be optionally attached to any of his/her relatives represented by the Relative class. It could be that the clinical data of any of the persons involved in this pedigree model exist elsewhere (in the same message or document, or in the person medical records). In this case it is possible to point to that data by including stub classes which only hold ids of the actual data, thus enabling applications to get the information if needed.
Person and Relative
The Person class allows the representation of personal information like gender and birth time of each of the patient's relatives. It is also linked back to the Relative class, an association which enables a recursive traversal a pedigree at any level of depth in a pure XML hierarchical fashion (i.e., only by nesting elements). Note that in each new traversal of this recursive association, the scoping Person represents another family member, and consequently the Relative class relates to this family member and not directly to the patient.
In general, the issue of recursion and XML hierarchy relate to the representation of pedigree data which is hierarchical by nature but could be represented in various ways using XML nesting elements on the one hand or mother and father ids for each relative thus constituting the hierarchy via links and pointers and not via nesting elements. The latter format also allows the flat outline of the pedigree. These issues are discussed in more detail in the documentation available from the HL7 Clinical Genomics SIG page in the HL7 site.
Relative - Mother and Father Identifiers
In a flat XML representation of a pedigree it is often required to maintain mother and father ids for each relative, in order to allow the family history application to reconstruct the pedigree. These ids should be placed in the id attribute of the Person class. That class should be populated for each parent if available. The first XML sample in appendix D shows a flat representation of a pedigree with the ids populated in the Person class (XML element is relationshipHolder.id).
It is required to use globally unique ids as mandated from the use of the II data type for the id attribute. Regarding the generation of thses ids, there can be two situations: (1) The application generating the pedigree has a 'root' OID dedicated to this purpose and it extends it for each pedigree it creates. Then, it generates arbitrary unique ids for each relative who doesn't have a globally unique id and places it in the extension component. The concatenation of the pedigree root and relative extension creates a globally unique id for the person; (2) The application knows a globally unique id of a relative and uses it. Note that in a certain pedigree there can be a mixture of the two situations.
| POCG_HD000040UV | POCG_HD000040UV01 |
|
||||||
|
For details on the interpretation of this section, see the description of HMDs in the Version 3 Guide.
| E_Organization | COCT_MT150000UV02 |
| A_GeneticLoci | COCT_MT540000UV |
| R_RelatedParty | COCT_MT910000UV |
| A_GeneticLocus | COCT_MT930000UV |
| Pedigree Event | POCG_MT000040UV01 |
|
||||||
|
For details on the interpretation of this section, see the definition of Interactions in the Version 3 Guide.
| Trigger Event | Original Pedigree Notification | POCG_TE000001UV01 |
| Transmission Wrapper | Send Message Payload | MCCI_MT000100UV01 |
| Control Act Wrapper | Trigger Event Control Act | MCAI_MT700201UV01 |
| Message Type | Pedigree Event | POCG_MT000040UV01 |
| Sender | Pedigree Sender | POCG_AR000001UV01 |
| Receiver | Pedigree Receiver | POCG_AR000002UV01 |
| Return to top of page |


