Clinical Genomics
 ANSI/HL7 V3 CGPED, R1-2007 (R2012) HL7 Version 3 Standard: Clinical Genomics; Pedigree, Release 1 (reaffirmation of ANSI/HL7 V3 CGPED, R1-2007) 6/21/2012 |
| Responsible Group | Clinical Genomics Work Group HL7 |
| Co-Chair, Facilitator & Primary Contributor | Amnon Shabo (Shvo), Ph.D. shabo@il.ibm.com IBM Research Lab in Haifa |
| Co-Chair | Kevin S. Hughes, M.D., FACS KSHUGHES@PARTNERS.ORG Massachusetts General Hospital, Partners HealthCare |
| Co-Chair | Mollie Ullman-Cullere MULLMANCULLERE@PARTNERS.ORG Harvard-Partners Center for Genetics and Genomics |
| Co-Chair | Scott Whyte scott.whyte@capgemini.com CGEY Health |
| Vocabulary Contributor | Usha Reddy, Ph.D. ushareddy@us.ibm.com IBM Life Sciences |
| Modeling & Vocabulary Contributor | Shosh Israel, Ph.D. israels@hadassah.org.il Hadassah University Hospital |
| Publishing Facilitator | Grant Wood grant.wood@intermountainmail.org Intermountain Healthcare Clinical Genetics Institute |
| Contributor and RCRIM liaison | Phil Pochon Phil.Pochon@covance.com Covance |
| Modeling and Vocabulary Contributor | Perry Mar PMAR@PARTNERS.ORG Partners HealthCare - Clinical Informatics Research and Development |
| Modeling & Vocabulary Contributor | Chuanbo Xu Chuanbo_Xu@perlegen.com Perlegen |
| MnM Facilitator (retired) | Charlie Mead, MD MSc Booz Allen Hamilton |
| Storyboard Contributor | Pravat Das Mayo Clinic |
| Storyboard Contributor | Jennifer Fostel, Ph.D. NIH - National Center for Toxicogenomics |
| Storyboard Contributor | Brent Gendleman 5AM Solutions |
| Storyboard Contributor | Rick Haddorff Mayo Clinic |
| Storyboard Contributor | Jim Holeman HP Nonstop Enterprise Division |
| Storyboard Contributor | Anajane Smith Fred Hutchinson Cancer Research Center |
| Storyboard Contributor | Derek Walker Fred Hutchinson Cancer Research Center |
| Storyboard Contributor | Sue-Jane Wang, Ph.D. US. Food and Drug Administration |
| Storyboard Contributor | Mathieu Wiepert Mayo Clinic |
| Storyboard Contributor | Katie Wittrup Mayo Clinic |
HTML Generated: 2012-08-31T12:06:56
Content Last Edited: 2012-02-24T15:12:20
HL7® Version 3 Standard, © 2007 Health Level Seven® International All Rights Reserved.
HL7 and Health Level Seven are registered trademarks of Health Level Seven International. Reg. U.S. Pat & TM Off.
Use of these materials is governed by HL7 International's IP Compliance Policy.
Table of Contents
Preface
i Notes to Readers
ii Message Design Element Navigation
1 Overview
1.1 Introduction & Scope
1.2 Domain Information Models
2 Pedigree Topic
2.1 Storyboards
2.2 Application Roles
2.3 Trigger Events
2.4 Refined Message Information Models
2.5 Hierarchical Message Descriptions
2.6 Interactions
3 Quality Analysis Report Topic
4 CMETs Defined by this Domain
5 CMETs Used by this Domain
6 Interactions Annex
6.1 By Application Role
6.2 By Trigger Event
6.3 By Message Type
7 Glossary
The Clinical Genomics WG has developed two topics - the Pedigree Topic (Family History), and the Genotype topic. The Pedigree Topic was approved by the membership as normative during the May 2007 ballot cycle and the final version is included in the Normative Edition beginning in 2008. The Genotype topic was approved as a DSTU, and subsequently updated, although the last DSTU period expired in 2010. The Clinical Genomics roadmap is to progress the DSTU to normative in a step-wise approach starting with an implementation guide for genetic variation. The latter can be found in the Clinical Genomics Domain currently under ballot. The reader is encouraged to use the implementation guide because it is a further constraining of the DSTU models and provide more elaborated and accurate description of how the models can be used to represent genetic variation data.
Since its formation, the Clinical Genomics SIG has been developing HL7 V3 standards to enable the exchange of interrelated clinical and personalized genomic data between interested parties. In many cases the exchange of genomic data is done between disparate organizations (healthcare providers, genetic labs, research facilities, etc.) and acceptable standards are crucial for the usefulness of genomic data in healthcare practice. It is envisioned that the use of genomic data in healthcare practice will become ubiquitous. Today, there are already several examples of the use of genomic data in healthcare and a few of them are presented in detail in the specifications' storyboards (e.g., Tissue Typing, Cystic Fibrosis, Pharmacogenomics and others use cases are presented briefly in the strategic direction of the SIG). These storyboards, unlike other ballot packages, are considered informative serving the purpose of illustrating the use of the Genotype models as the basic units of genomic data encapsulation. Indeed, the Genotype models (GeneticLocus and GeneticLoci) can be utilized by any group in HL7 that needs to convey genomic data.
The core Genotype model is the GeneticLocus model. It consists of various types of genomic data relating to a specific DNA locus including sequencing, expression and proteomic data. Within the GeneticLocus model we have utilized existing bioinformatics markups to represent raw data received from genomic facilities. Examining and constraining these markups is a work in progress and thus this part of the GeneticLocus model is considered informative as well.
The FamilyHistory model is aimed at describing a patient's pedigree with genomic data. Note that this model utilizes the GeneticLocus and GeneticLoci models (as CMETs) to carry the genomic data for the patient's relatives. The requirement for an elaborated Clinical Genomic Family History model has been identified while working on the BRCA (Breast Cancer) Storyboard. The Family History storyboard includes parts of the full BRCA storyboard as examples of how one might use the Family History model and interaction set.
|
The Clinical Genomics domain addresses requirements for the interrelation of clinical and genomic data at the individual level. Much of the genomic data is still generic, for example the human genome is in fact the DNA sequences believed to be the common sequences in every human being. The vision of 'personalized medicine' is based on those correlations that make use of personal genomic data such as the SNPs (Single Nucleotide Polymorphisms) that differentiate any two persons and occur about every thousand bases. Beside normal differences, health conditions such as drug sensitivities, allergies and others could be attributed to the individual SNPs or to differences in gene expression and proteomics.
The emphases the Clinical Genomics domain are the personalization of the genomic data and the 'intelligent' linking to relevant clinical information. These links are probably the main source from which geneticists (genomicists?) and clinicians could benefit. The cases where genomic data are used in healthcare practice vary in complexity and extent of the data used, since the current testing methods are still very expensive and not widely used. We can see simple testing like identifying genes and mutations as well as full sequencing of alleles and the use of micro-arrays to identify the expression of vast number of genes in each individual. Naturally, the Clinical Genomics SIG has been focusing on tests that are routinely done in healthcare, while preparing the information infrastructure standard for more futuristic cases.
At a first sight it seems that genomic data sets are yet another type of observations. While this is true of course, there are a few characteristics that might distinguish it from typical observations such as blood pressure or potassium level:
-
The amount of data: potentially it could be the entire human genome
-
The personalization of the data is evolving as new discoveries are constantly made
-
The complexity of the data: not only the DNA sequences (...AGCT...) need to be represented, but also SNPs, annotations (automatic and manual), gene expression, protein translation, and more
-
The emerging standard formats being used by bioinformatics communities, for example: BSML (Bioinformatic Sequence Markup Language), MAGE-ML ( Microarray and GeneExpression Markup Language)
-
Various standard organizations and many stakeholders are involved
-
The clinical-genomic correlations are represented in variety of different ways depending on the point of view (clinical research, pharmaceutical or healthcare)
The Clinical-Genomics SIG develops HL7 standards to enable the exchange between interested parties of the clinical and personalized genomic data. In many case the exchange of genomic data is done between disparate organization (providers, labs, research facilities, etc.) and acceptable standards are crucial for the usefulness of the data in healthcare practice.
Diagram
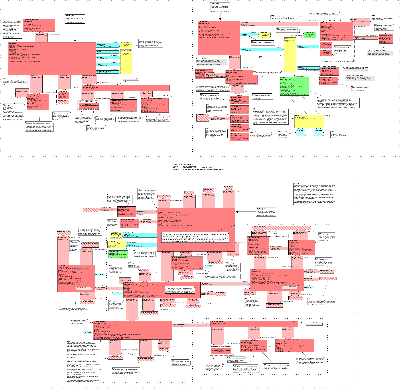
The Clinical Genomics DIM is the result of our effort to look for the commonalities in each genomic-oriented storyboard that we have been working on (i.e., Tissue Typing, Cystic Fibrosis, BRCA and Pharmacogenomics). The first model we developed was called the Genotype model and it described a single locus on chromosomal or mitochondrial DNA. We then developed a model to describe a set of genetic loci and we now have two core models within the Genotype Topic: GeneticLocus and GeneticLoci. In parallel, we developed a Clinical Genomics Family History model. The DIM is an integration of these three models. For deatiled description of each class in the DIM, please refer to the respective walk through of each of the above-mentioned models. The trial use of this draft standard by its early adopters will guide the crystallization of this DIM into a more generic model for this domain.
In this ballot cycle we present two new models for comments only: (1) a genetic test order model is introduced at the end of the GeneticLoci model description (since it uses the GeneticLoci model as a CMET) and (2) a query model which is presented below.
The Clinical Genomics Query Model:
General:
HL7 V3 has a framework for domain-specific query models described in the following path of the V3 ballot package: Specifications Infrastructure / Messaging / Query Infrastructure. It states that the Query Infrastructure domain specifies the formation of information queries and the responses to these queries to meet the needs of healthcare applications using the HL7 version 3 messaging standard. In addition, the Query Infrastructure domain asserts that the variety of potential queries is almost unlimited and HL7 does not try to cover every possible query. Rather, the Query Standard provides general methods by which to structure query/response pairs. The technical committees responsible for functional domains apply these methods to develop specific query/response pairs for those domains' needs.
The Clinical Genomics Query Model:
The Clinical Genomics SIG develops a Query model based on the Query Infrastructure described above. It model is presented for the first time in the January 2007 ballot cycle - for comments only. The model is based on a query-by-parameter approach where the domain groups pick attributes in their domain and declares them to be parameters of the domain query, while constraining them if necessary. We picked a few central attributed from the GeneticLocus and GeneticLoci models and placed them as parameters in the Clinical Genomics Query model. The development is done step-wise and in the future we'll add parameters from the GeneticTestOrder model as well as the FamilyHistory model.
Query Interaction:
The basic interaction where a query message could be used is as follows: an application sends a query message to another application where certain parameters have been populated with data values. The receiving application parses the query message and uses the parameters with their assigned values to query its repository of HL7 Clinical Genomics messages or any other repository where there is a clear mapping between its data model and the HL7 Clinical Genomics models. All messages that meet the criteria set by the parameters and values will be returned to the sending application. Thus, the response is always a set of zero to many valid HL7 instances.
Note that an HL7 interaction where this message can be used has not been developed yet. So the model presented here for comments only is a static model. Nevertheless, it's encouraged to experiment with this model and simulate a query scenario in clinical genomics and see if at all it can meet the requirements or perhaps this query paradigm is too simplistic for the clinical genomics domain.
The figure below shows the Query model.
| Return to top of page |